https://doi.org/10.3389/fgene.2023.1108416
Abstract
Sesame is an ancient oilseed crop containing many valuable nutritional components. The demand for sesame seeds and their products has recently increased worldwide, making it necessary to enhance the development of high-yielding cultivars. One approach to enhance genetic gain in breeding programs is genomic selection. However, studies on genomic selection and genomic prediction in sesame have yet to be conducted.
In this study, we performed genomic prediction for agronomic traits using the phenotypes and genotypes of a sesame diversity panel grown under Mediterranean climatic conditions over two growing seasons. We aimed to assess prediction accuracy for nine important agronomic traits in sesame using single- and multi-environment analyses.
In single-environment analysis, genomic best linear unbiased prediction, BayesB, BayesC, and reproducing kernel Hilbert spaces models showed no substantial differences. The average prediction accuracy of the nine traits across these models ranged from 0.39 to 0.79 for both growing seasons. In the multi-environment analysis, the marker-by-environment interaction model, which decomposed the marker effects into components shared across environments and environment-specific deviations, improved the prediction accuracies for all traits by 15%–58% compared to the single-environment model, particularly when borrowing information from other environments was made possible.
Our results showed that single-environment analysis produced moderate-to-high genomic prediction accuracy for agronomic traits in sesame. The multi-environment analysis further enhanced this accuracy by exploiting marker-by-environment interaction. We concluded that genomic prediction using multi-environmental trial data could improve efforts for breeding cultivars adapted to the semi-arid Mediterranean climate.
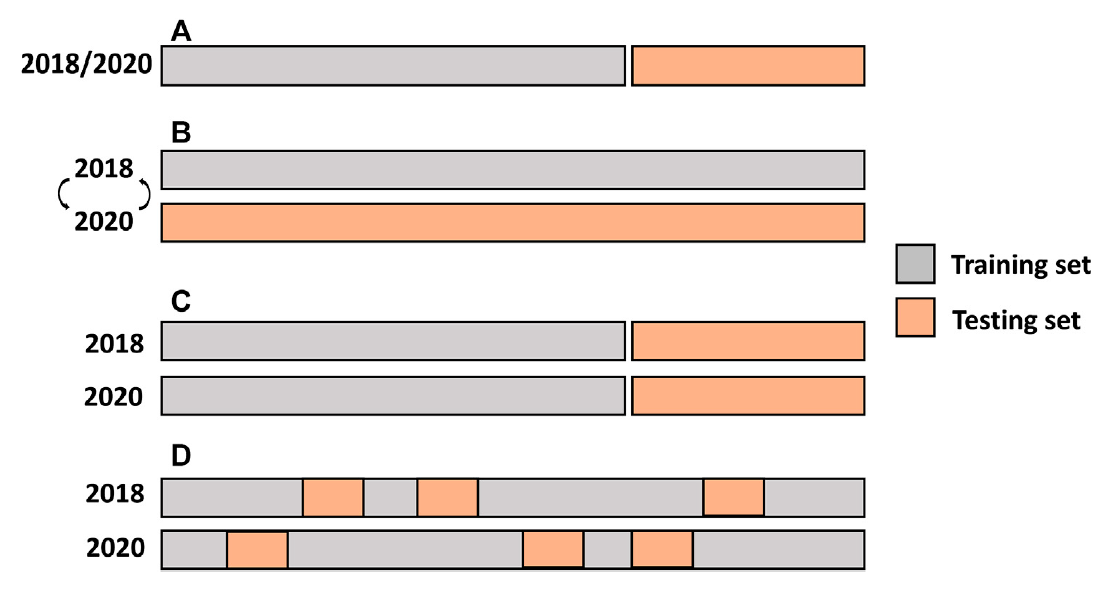
FIGURE 1. Single-and multi-environment genomic prediction cross-validation scenarios. (A) Single-environment analysis, (B) All the lines in one environment were used to predict the same lines in a new environment (CV0), (C) Performance of new lines that are not phenotyped in any environment was predicted through the genetic relationship with other lines (CV1), and (D) Predict lines that were evaluated in only one environment through the genetic and environmental relationships (CV2).
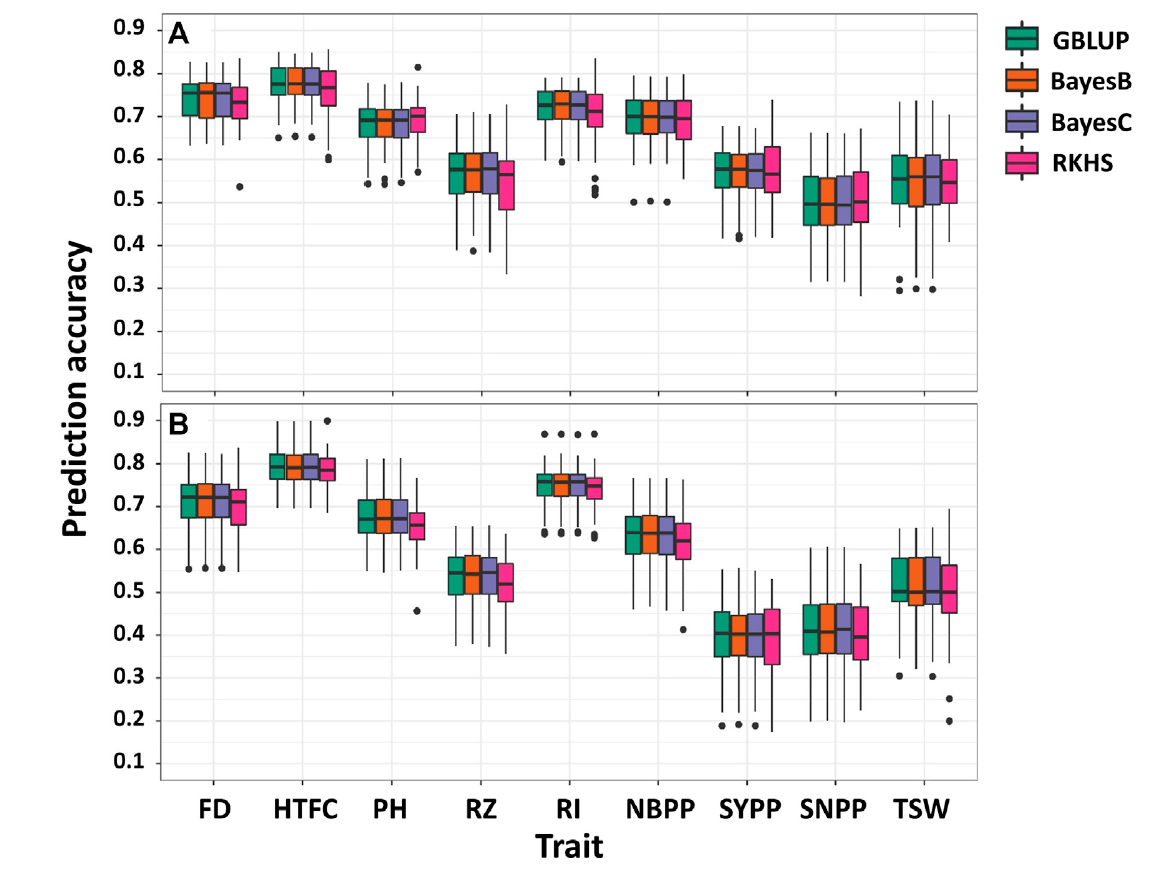
FIGURE 2. Single-environment prediction accuracies of the nine agronomic sesame traits in 2018 (A) and 2020 (B) growing seasons using genomic best linear unbiased prediction (GBLUP), BayesB, BayesC, and reproducing kernel Hilbert spaces regression (RKHS). Flowering date (FD), height to the first capsule (HTFC), plant height (PH), reproductive zone (RZ), reproductive index (RI), number of branches per plant (NBPP), seed-yield per plant (SYPP), seeds number per plant (SNPP), and thousand-seed weight (TSW).
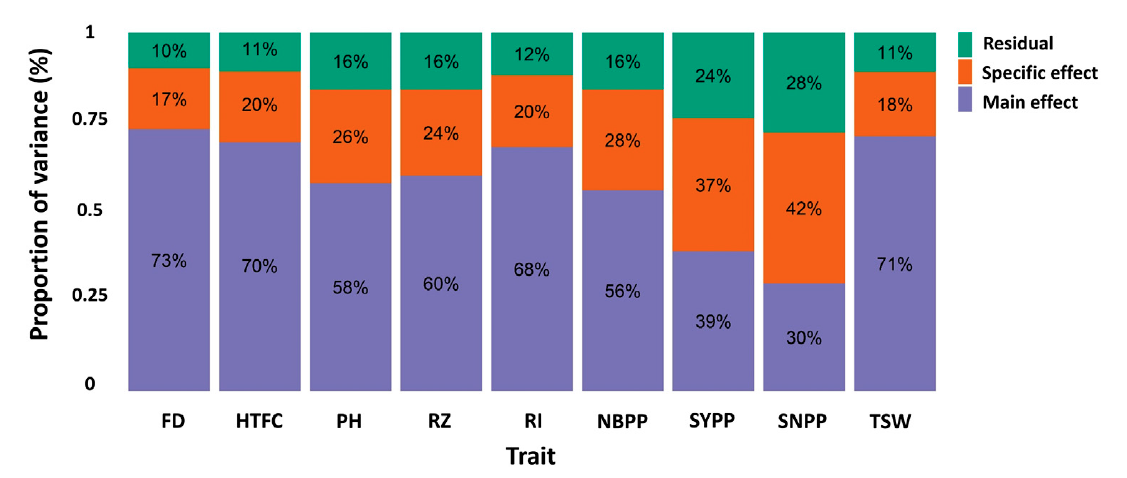
FIGURE 3. Proportion of the main genetic variance (main effect), environment-specific variance (specific effect), and residual variance components for each trait obtained from the marker-by-environment interaction model. Flowering date (FD), height to the first capsule (HTFC), plant height (PH), reproductive zone (RZ), reproductive index (RI), number of branches per plant (NBPP), seed-yield per plant (SYPP), seeds number per plant (SNPP), and thousand-seed weight (TSW).
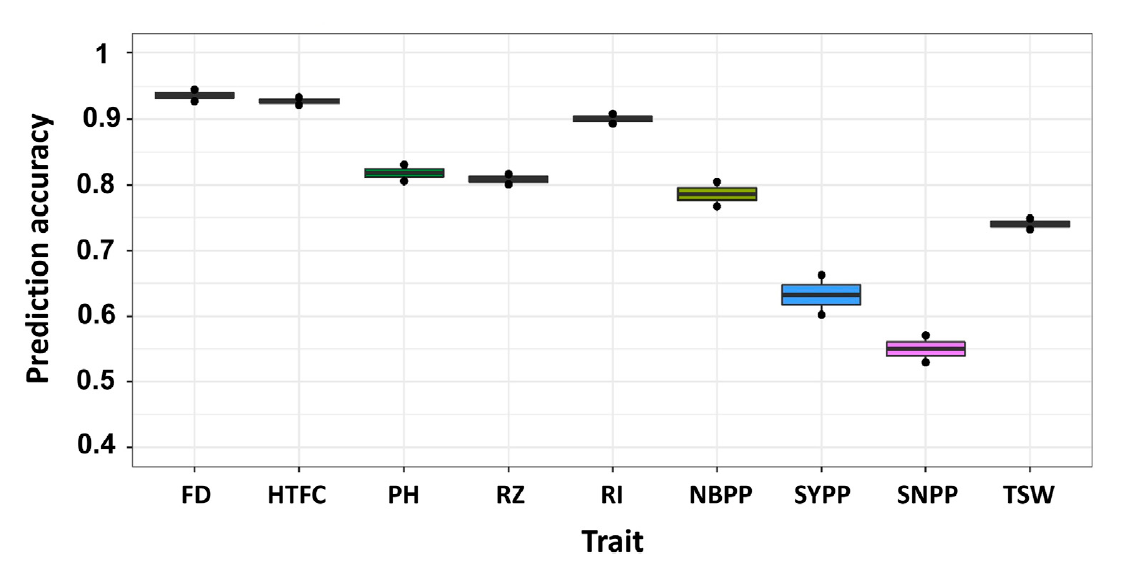
FIGURE 4. Multi-environment genomic prediction accuracies of the nine agronomic sesame traits using the best linear unbiased prediction model when all the lines in one environment were used to predict the same lines in a new environment (CV0). Flowering date (FD), height to the first capsule (HTFC), plant height (PH), reproductive zone (RZ), reproductive index (RI), number of branches per plant (NBPP), seed-yield per plant (SYPP), seeds number per plant (SNPP), and thousand-seed weight (TSW).
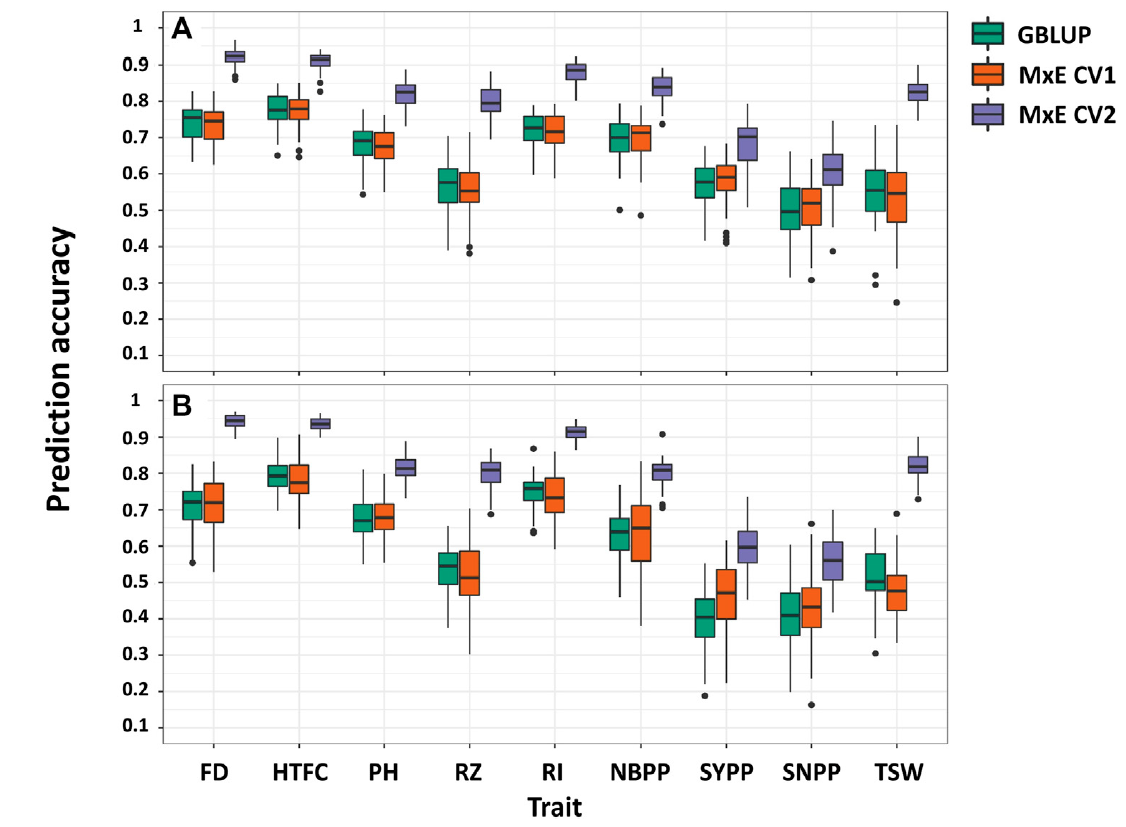
FIGURE 5. Comparison of prediction accuracies in single- and multi-environment models for predicting new lines that are not phenotyped in any environment (CV1) and predicting lines that were evaluated in only one environment (CV2) in 2018 (A) and 2020 (B) growing seasons. Flowering date (FD), height to the first capsule (HTFC), plant height (PH), reproductive zone (RZ), reproductive index (RI), number of branches per plant (NBPP), seed-yield per plant (SYPP), seeds number per plant (SNPP), and thousand-seed weight (TSW).