Paper: https://doi.org/10.1093/g3journal/jkad052
Codes: https://github.com/yebigithub/VTUNL_Rice
Abstract
The asymmetric increase in average nighttime temperatures relative to increase in average daytime temperatures due to climate change is decreasing grain yield and quality in rice. Therefore, a better genome-level understanding of the impact of higher night temperature stress on the weight of individual grains is essential for future development of more resilient rice. We investigated the utility of metabolites obtained from grains to classify high night temperature (HNT) conditions of genotypes, and metabolites and single-nucleotide polymorphisms (SNPs) to predict grain length, width, and perimeter phenotypes using a rice diversity panel. We found that the metabolic profiles of rice genotypes alone could be used to classify control and HNT conditions with high accuracy using random forest or extreme gradient boosting. Best linear unbiased prediction and BayesC showed greater metabolic prediction performance than machine learning models for grain-size phenotypes. Metabolic prediction was most effective for grain width, resulting in the highest prediction performance. Genomic prediction performed better than metabolic prediction. Integrating metabolites and genomics simultaneously in a prediction model slightly improved prediction performance. We did not observe a difference in prediction between the control and HNT conditions. Several metabolites were identified as auxiliary phenotypes that could be used to enhance the multi-trait genomic prediction of grain-size phenotypes. Our results showed that, in addition to SNPs, metabolites collected from grains offer rich information to perform predictive analyses, including classification modeling of HNT responses and regression modeling of grain-size-related phenotypes in rice.
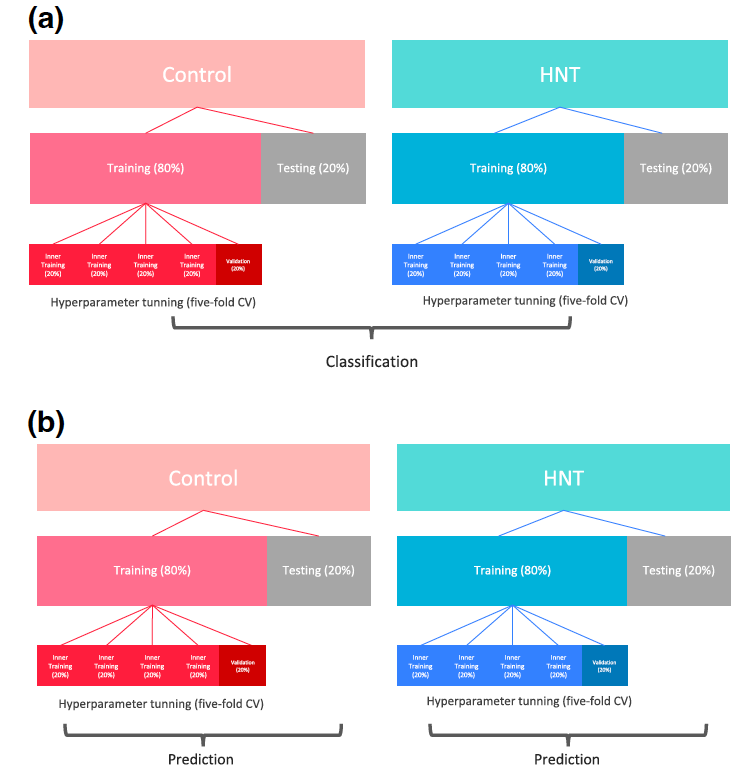
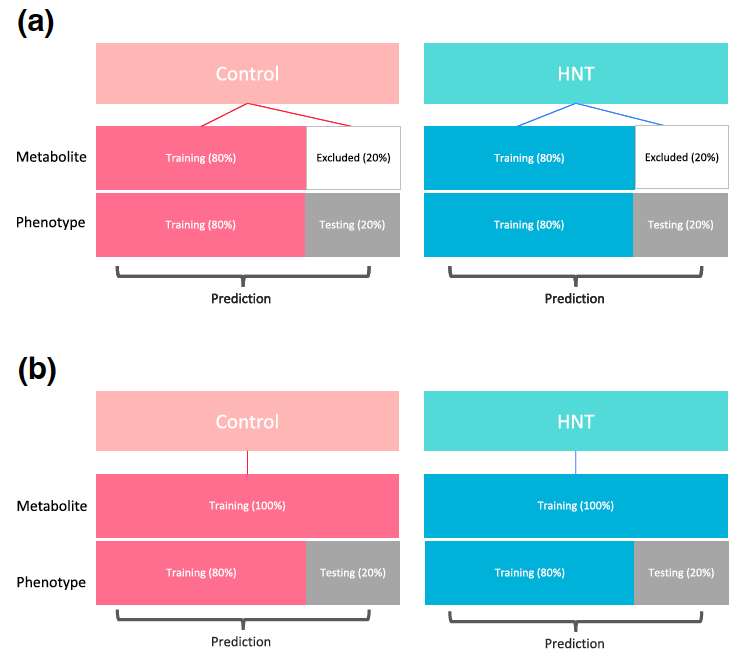
Figure 1: Cross-validation (CV) design for binary classification of high night temperature stress conditions (A) and metabolimic and genomic prediction of grain size related pheno- types (B). Figure 2: Cross-validation design for multi-trait prediction. Scenario 1 (A) and Scenario 2 (B).
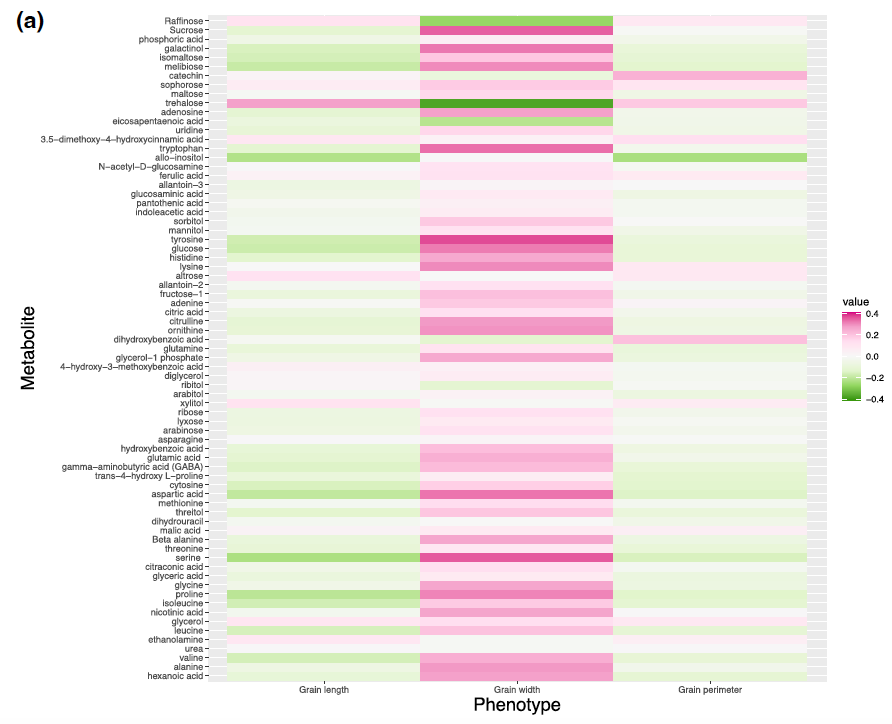
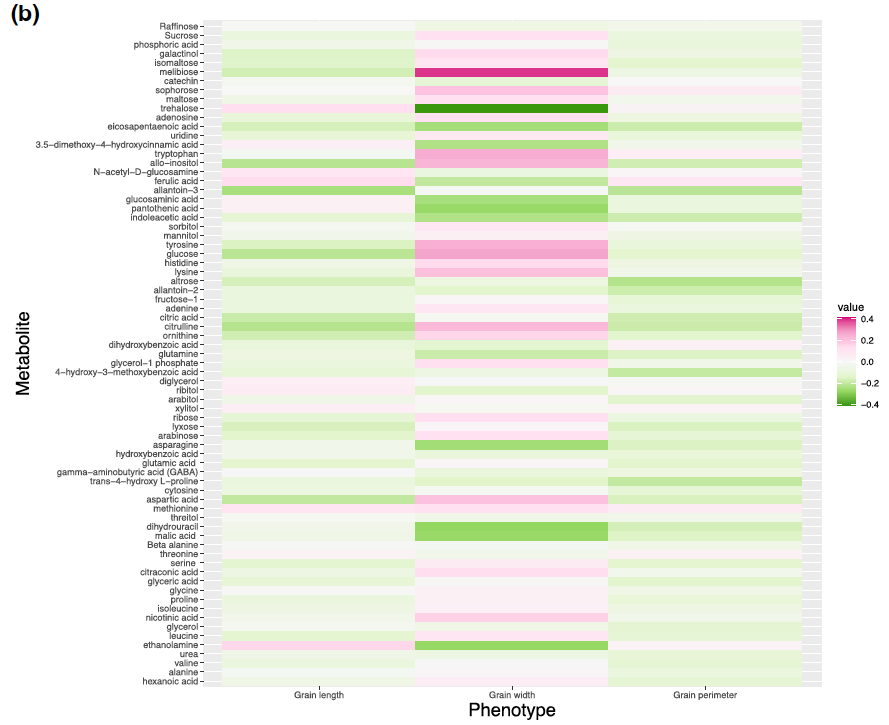
Figure 3: Correlation between metabolites and grain size phenotypes in control (A) and high night time temperature stress (B) conditions.
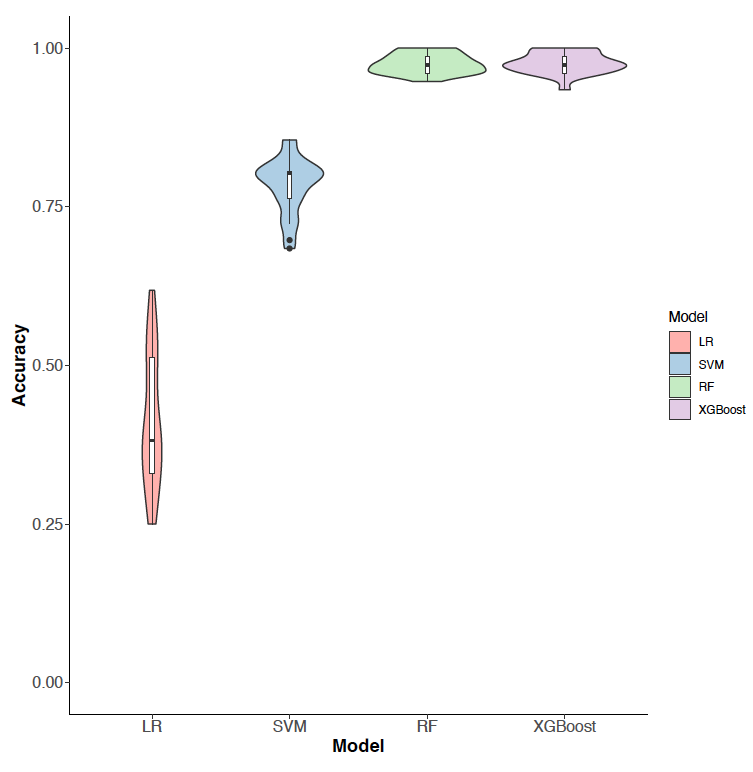
Figure 4: Classification accuracy of high night time temperature conditions (control and stress) using 73 metabolites. LR: logistic regression; SVM: support vector machine; RF: random forest; and XGBoost: extreme gradient boosting.
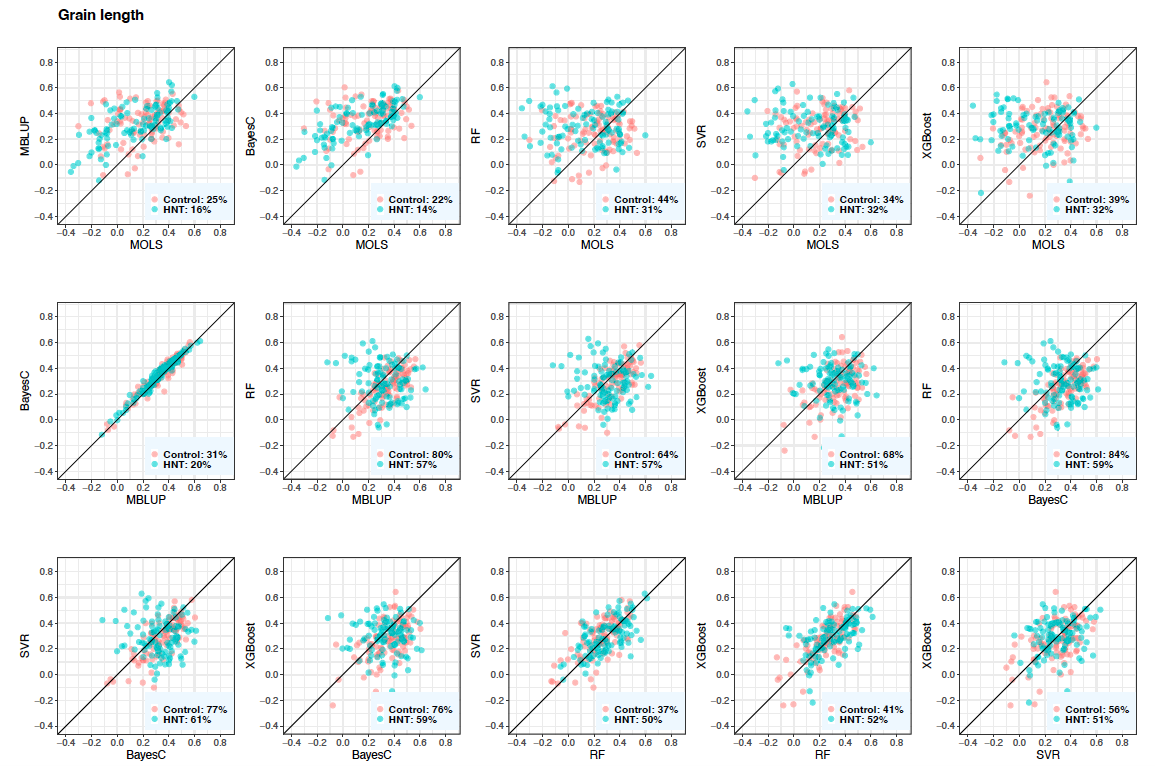
Figure 5: Predictive correlations of grain length using metabolic prediction in control and high night time temperature stress conditions. The percentages on the bottom right show the number of cross-validation resampling runs that the model on the x-axis performed better than the model on the y-axis. MOLS: metabolic ordinary least squares; MBLUP: metabolic best linear unbiased prediction; RF: random forests; SVR: support vector regression; and XGBoost: extreme gradient boosting.
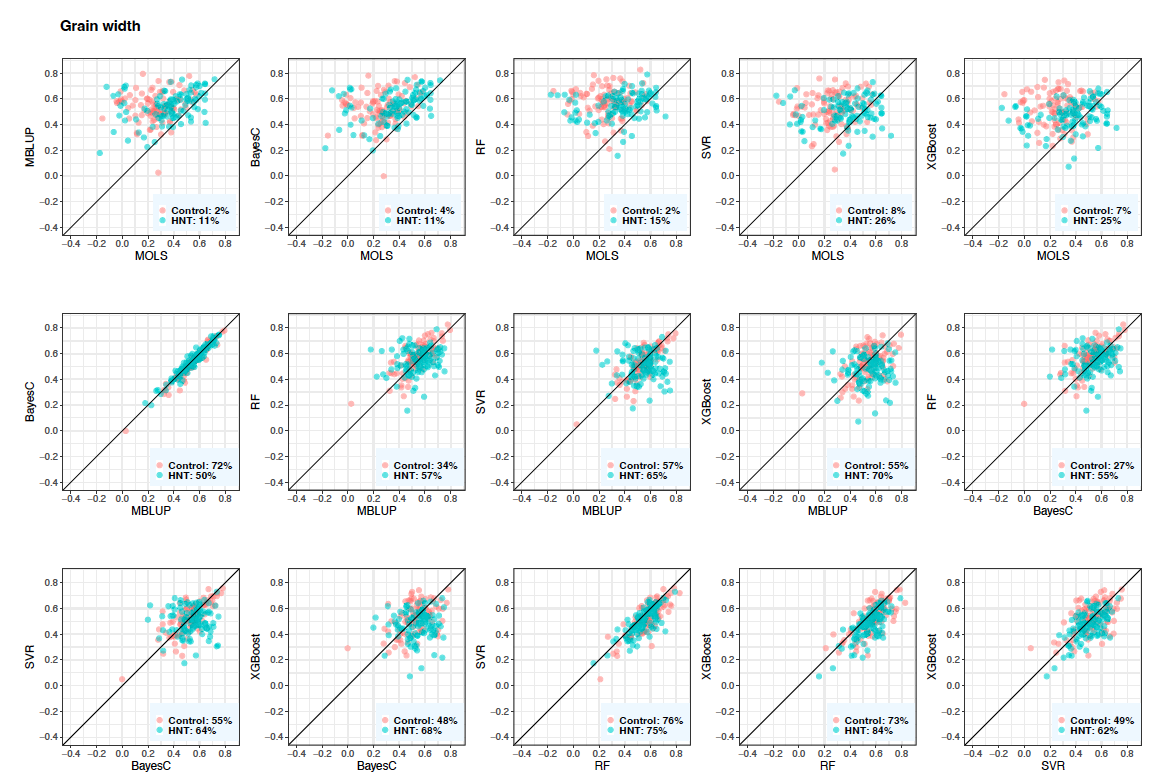
Figure 6: Predictive correlations of grain width using metabolic prediction in control and high night time temperature stress conditions. The percentages on the bottom right show the number of cross-validation resampling runs that the model on the x-axis performed better than the model on the y-axis. MOLS: metabolic ordinary least squares; MBLUP: metabolic best linear unbiased prediction; RF: random forests; SVR: support vector regression; and XGBoost: extreme gradient boosting.
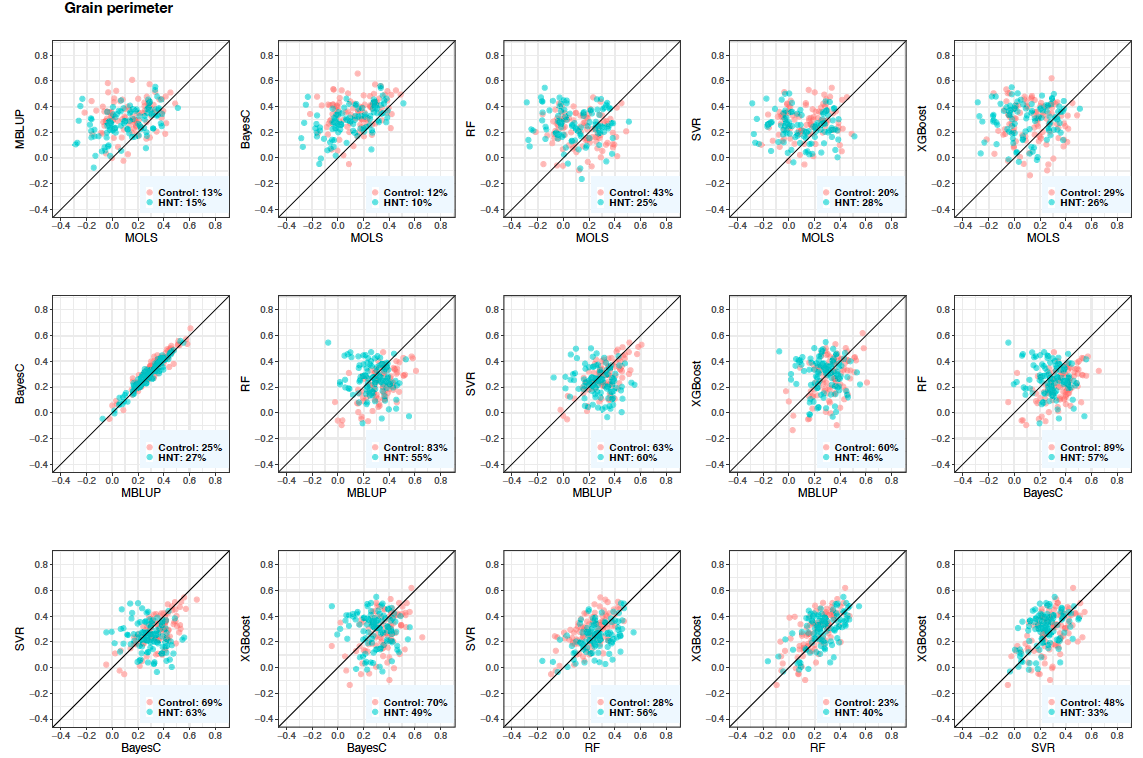
Figure 7: Predictive correlations of grain perimeter using metabolic prediction in control and high night time temperature stress conditions. The percentages on the bottom right show the number of cross-validation resampling runs that the model on the x-axis performed better than the model on the y-axis. MOLS: metabolic ordinary least squares; MBLUP: metabolic best linear unbiased prediction; RF: random forests; SVR: support vector regression; and XGBoost: extreme gradient boosting.
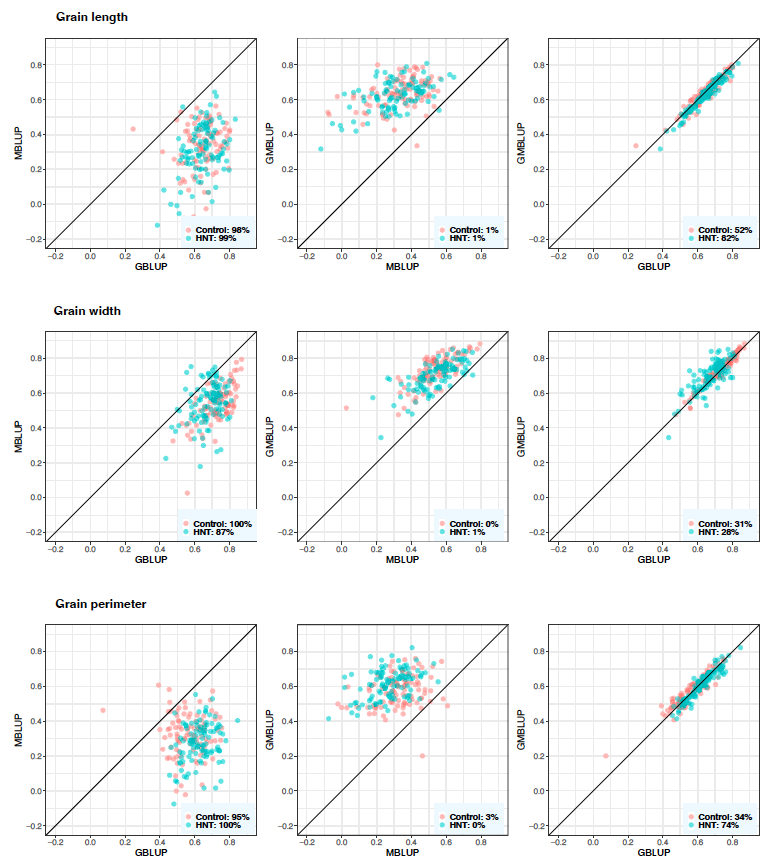
Figure 8: Predictive correlations of grain length, grain width, and grain perimeter using metabolic, genomic, and multi-omic prediction models in control and high night time tem- perature stress conditions. The percentages on the bottom right show the number of cross- validation resampling runs that the model on the x-axis performed better than the model on the y-axis. MBLUP: metabolic best linear unbiased prediction; GBLUP: genomic best linear unbiased prediction; and GMBLUP: genomic and metabolic best linear unbiased prediction
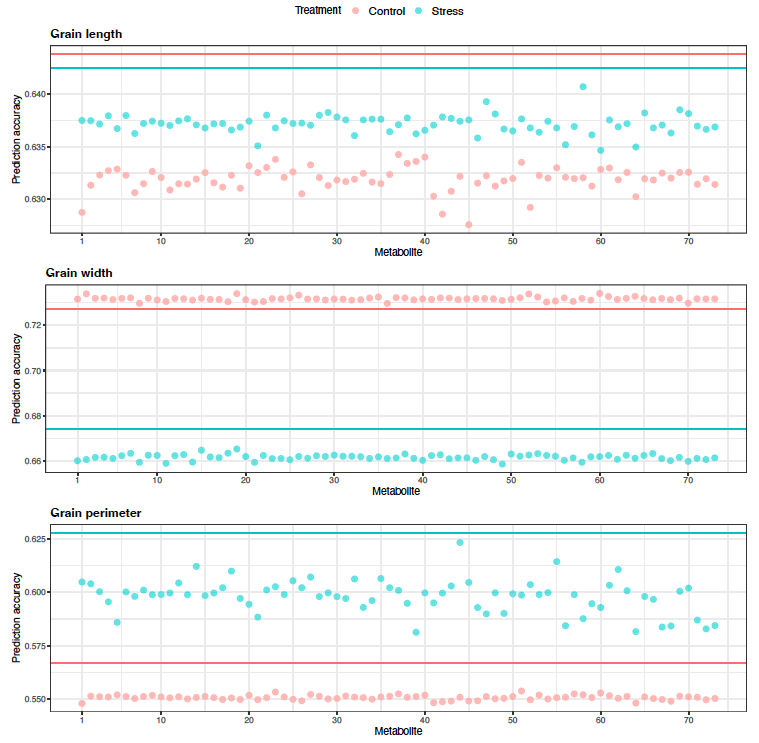
Figure 9: Predictive correlations of Scenario 1 multi-trait (bivariate) genomic prediction for grain length, grain width, and grain perimeter when metabolites were used as a secondary phenotype under control and high night time temperature stress conditions. The horizontal lines indicate the predictive correlations of single-trait genomic prediction.
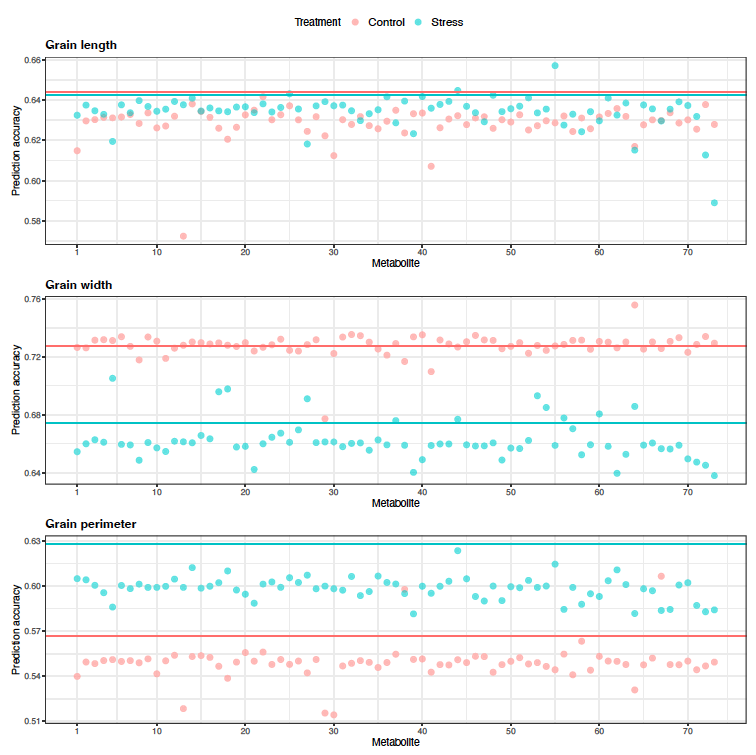
Figure 10: Predictive correlations of Scenario 2 multi-trait (bivariate) genomic prediction for grain length, grain width, and grain perimeter when metabolites were used as a secondary phenotype under control and high night time temperature stress conditions. The horizontal lines indicate the predictive correlations of single-trait genomic prediction.